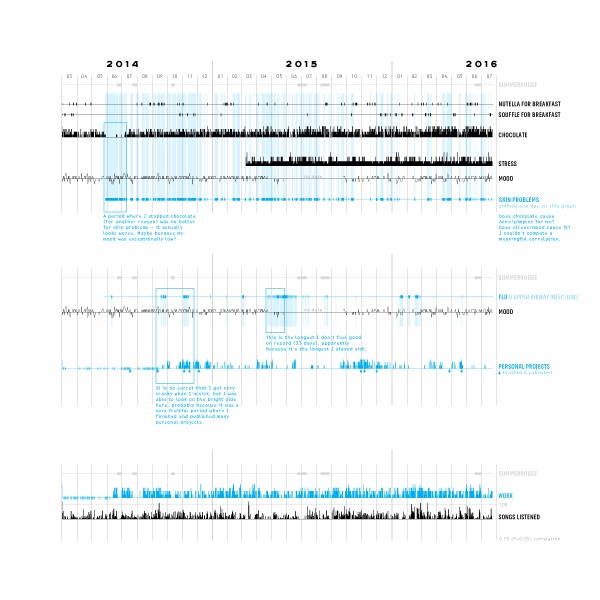
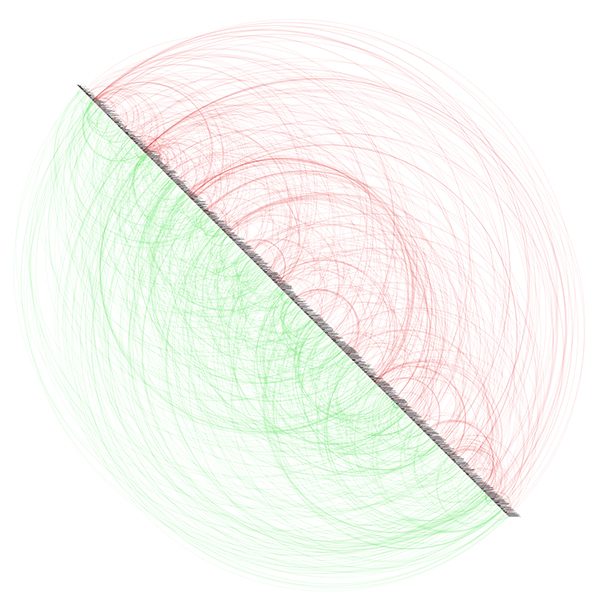
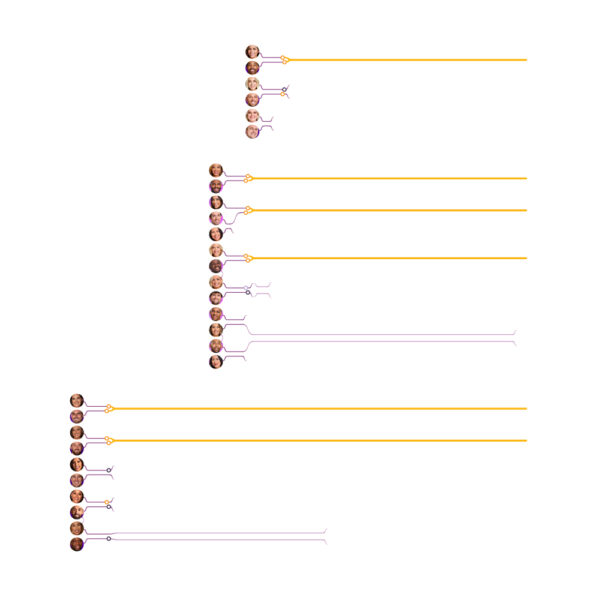
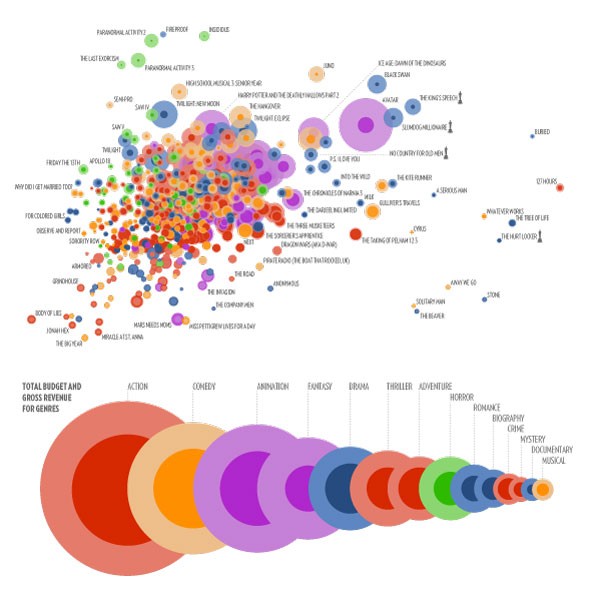
Visualizing Protein Motions
I worked with Canan Atılgan and Ali Rana Atılgan on the figures of their paper “Network-based Models as Tools Hinting at Non-evident Protein Functionality” in Annual Review of Biophysics, Vol. 41. There, I once again faced the challenge of showing protein dynamics in static images; since scientists usually publish in static (printed) media, showing these motions in figures is a common problem because proteins are extremely complex structures and even drawing them without motion is a nightmare.
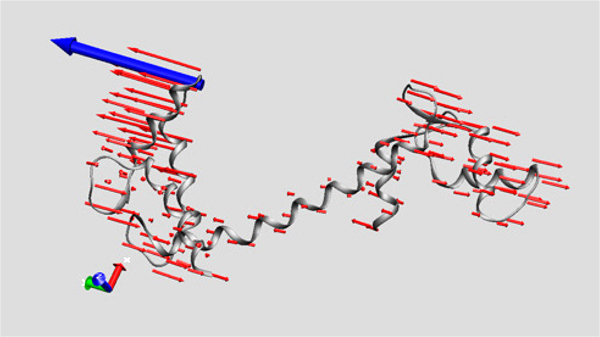 I had had some previous experience with this problem in Image of Science 2010 in Ayşe Özlem Aykut’s project, and then in the Structural Biology class I took in Sabancı University. The method I saw being used was putting in lots of red arrows/cones (one for each residue), and I wasn’t happy with how they looked; ugly, busy, and uninformative.
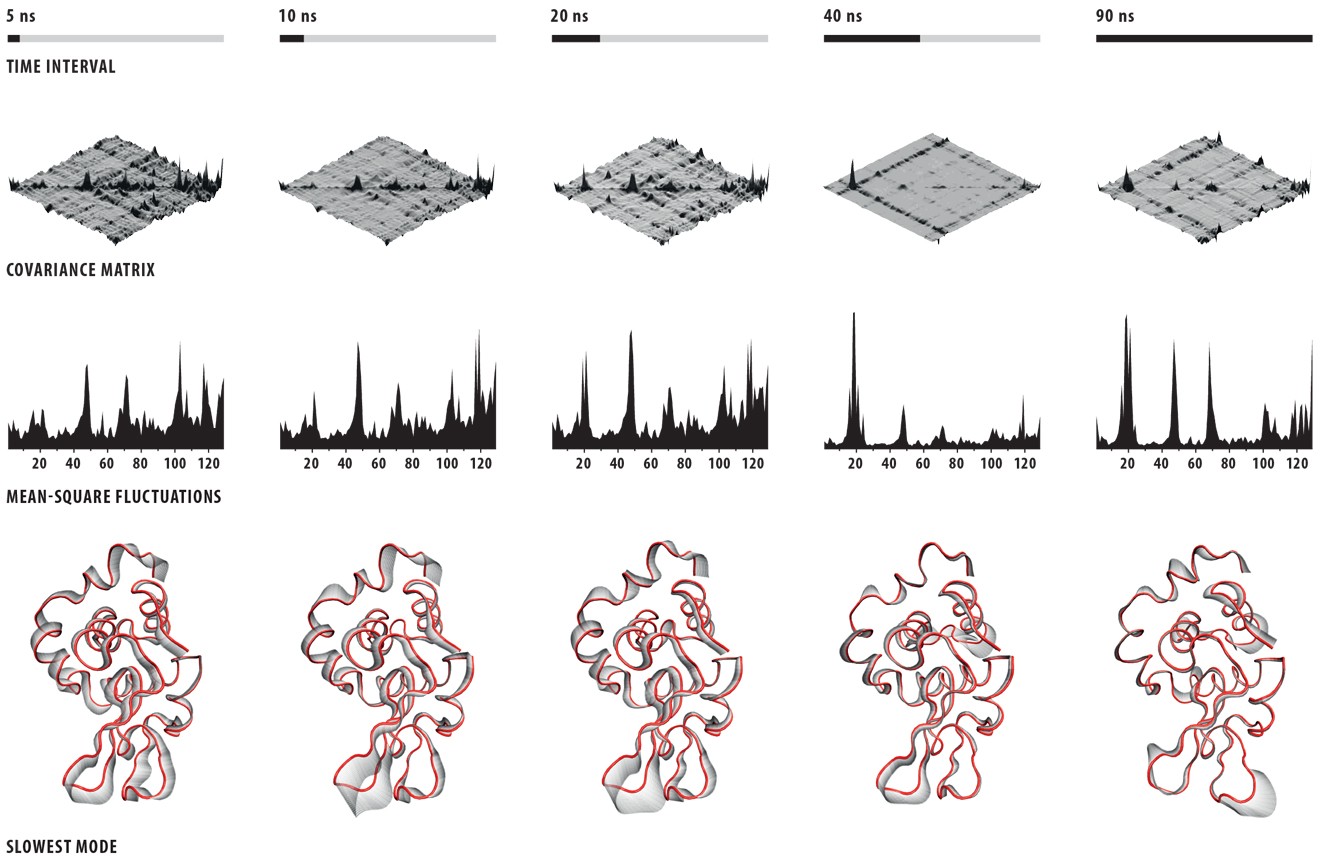
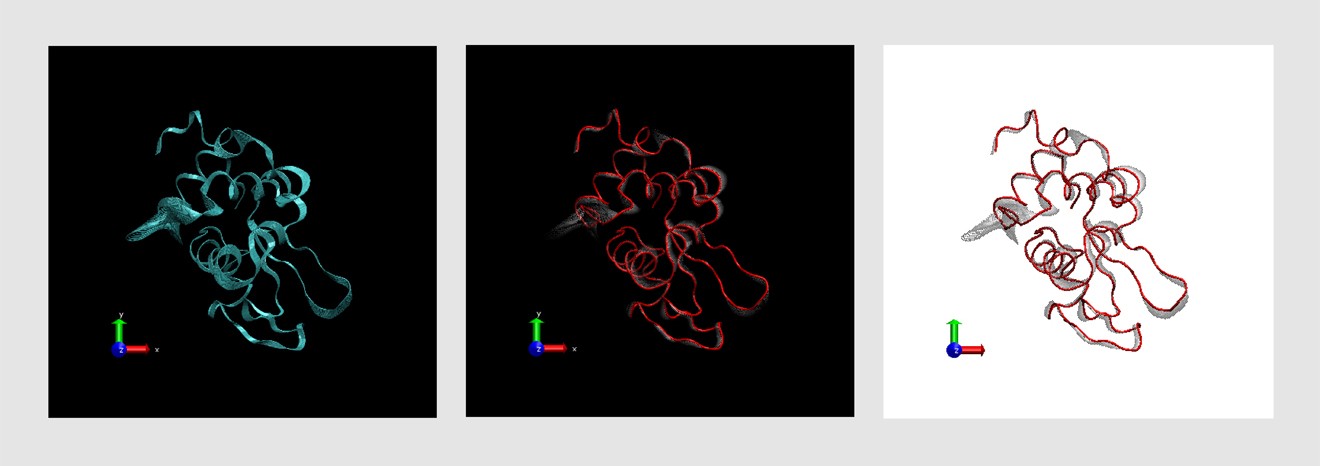
I had had some previous experience with this problem in Image of Science 2010 in Ayşe Özlem Aykut’s project, and then in the Structural Biology class I took in Sabancı University. The method I saw being used was putting in lots of red arrows/cones (one for each residue), and I wasn’t happy with how they looked; ugly, busy, and uninformative.This time Canan and I worked out a method (within the software VMD – Visual Molecular Dynamics) that we thought was more successful (and good-looking) than the arrows in showing which parts move to what degree and in which direction. Below is the figure I prepared for that paper, and at the bottom of the figure you can see the motion visualization that we created.
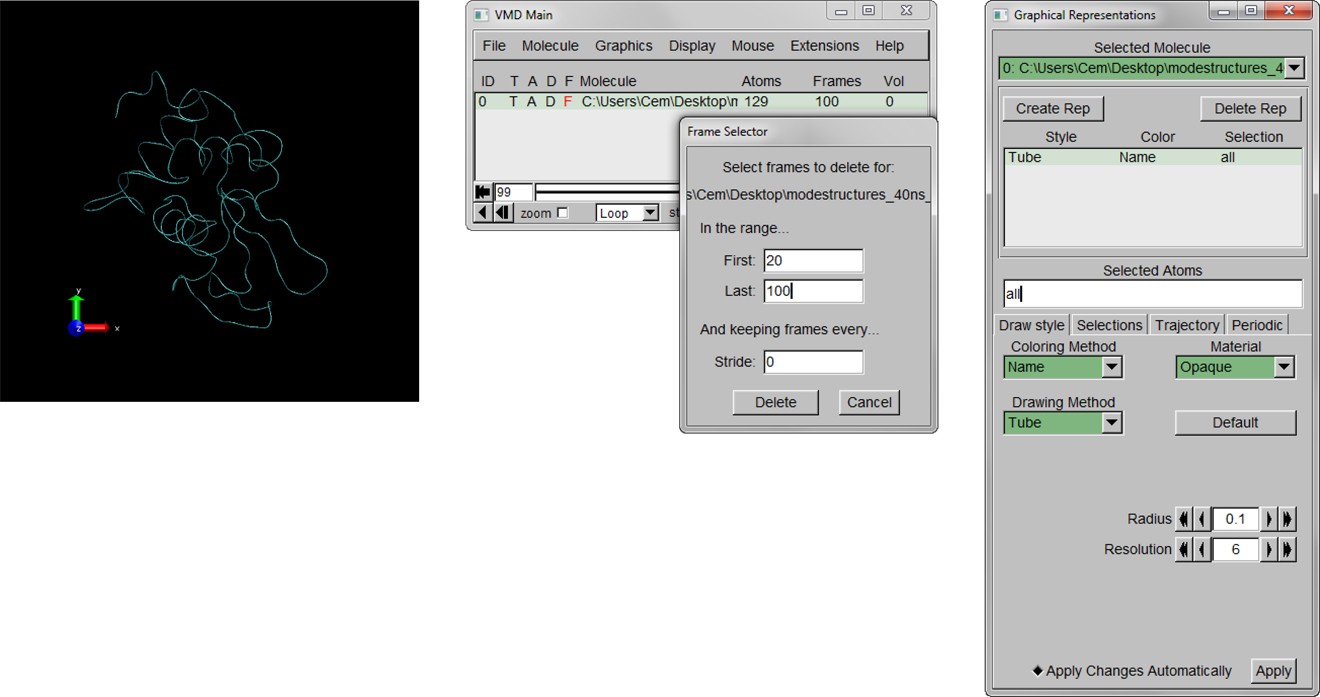
Here is the detailed tutorial for those who want to use this method.
Here is the detailed tutorial for those who want to use this method.